- Details

Cytomics Reactome. The trillions of cells in the adult human body, specialized to fulfill diverse roles in tissues, organs, and organ systems, all originate from a single cell, a zygote formed at conception. From zygote to adulthood, cells divide and commit to different fates. The steps from a pluripotent stem cell to a specialized descendant constitute a cell lineage path. Cytomics Reactome extends the data structure used for molecular-level annotation of cell biology to annotate cell lineage paths, organizing them by organ systems, cross-referencing them to Gene Ontology biological processes, and dividing each into causally connected cell development steps. These reaction-like steps describe transitions between cell states during development or differentiation, characterized by positive and negative regulators and required input components (cell state biomarkers required for the action of regulators). Each cell state corresponds to a unique combination of cell type (Cell Ontology), anatomical location (UBERON), and protein and/or RNA markers, as described in the Cytomics section of the User Guide. Recently developed tools that harvest omics data from single cells of multicellular organisms and track fates open the door to deciphering cell lineage paths at single-cell resolution, a critical requirement of regenerative medicine and cancer medicine. Cytomics Reactome is intended to support such technologies and their application to human biology as they emerge.
The first Cytomics cell lineage path, Differentiation of keratinocytes in interfollicular epidermis in mammalian skin, describes the differentiation of keratinocytes from stem cells to corneocytes in the interfollicular epidermis, the skin surface layer in between the adnexa (hair follicles, sweat glands, and sebaceous glands). The path is described in four cell differentiation steps involving five distinct cellular states: keratinocyte stem cells of the epidermal basal layer, transit amplifying cells, spinous keratinocytes, granular keratinocytes, and corneocytes. Each differentiation step is regulated by a distinct combination of regulatory molecules present in the microenvironments of the differentiating cells.
New and Updated Topics and Pathways. Topics with new or revised pathways in this release include Cell Cycle (Resolution of Sister Chromatid Cohesion), Developmental Biology (Differentiation of keratinocytes in intefrollicular skin epidermis, MITF-M-regulation melanocyte development, Transcriptional regulation of brown and beige adipocyte differentiation), Disease (Defective regulation of TLR by endogenous ligand, Signaling by LTK in cancer), Immune System (Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE), Regulation of TLR by endogenous ligand, TICAM1-dependent activation of IRF3/IRF7 ), Metabolism (Acetylcholine regulates insulin secretion, Adrenaline,noradrenaline inhibits insulin secretion, Aerobic respiration and respiratory electron transport, AMPK inhibits chREBP transcriptional activation activity, Branched-chain amino acid catabolism, ChREBP activates metabolic gene expression, Citric acid cycle (TCA cycle), Glucagon signaling in metabolic regulation, Glucagon-like Peptide-1 (GLP1) regulates insulin secretion, Heme biosynthesis, Malate-aspartate shuttle, Maturation of TCA enzymes and regulation of TCA cycle, NADPH regeneration, PI and PC transport between ER and Golgi membranes, PKA-mediated phosphorylation of key metabolic factors, Pyruvate metabolism, Regulation of pyruvate dehydrogenase (PDH) complex, Respiratory electron transport), Metabolism of proteins (Protein lipoylation), Signal Transduction (EGFR Transactivation by Gastrin, Signaling by LTK), Transport of small molecules (Iron uptake and transport).
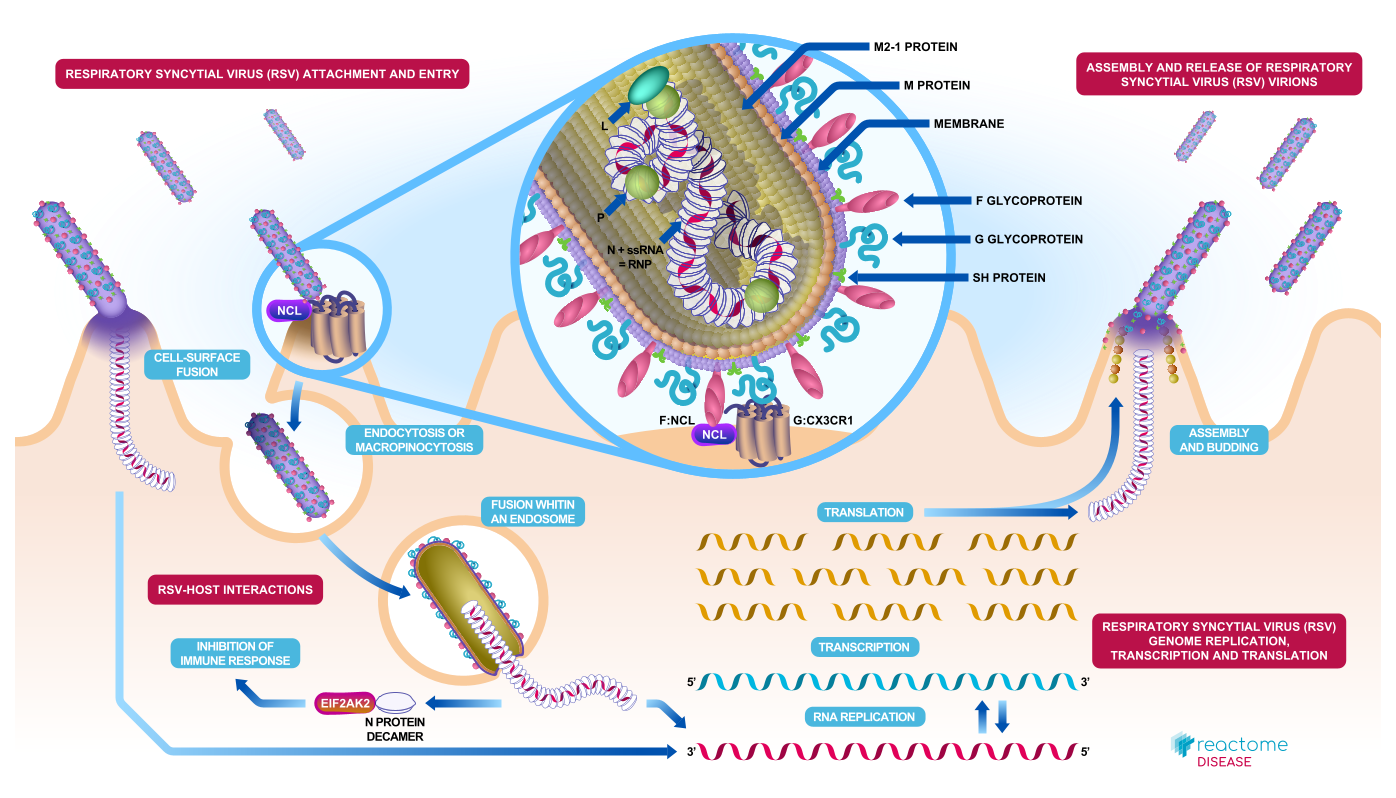
New and Updated Illustrations. New or revised Illustrations with embedded navigation features have been created for Adipogenesis, Developmental Biology, Developmental Cell Lineages, Diseases associated with TLR signaling cascade, Diseases of signal transduction by growth factor receptors and second messengers, Metabolism, MITF-M-regulated melanocyte development, Post-translational protein modification, and Signalling by Receptor Tyrosine Kinases. A new static illustration is available for Respiratory syncytial virus (RSV) attachment and entry.
Thanks to our Contributors. Tae-Hwa Chun, Toni Franjkić, Clarisse Ganier, Heinz Arnheiter, David Hill, Xing Liu, Ivana Munitic, and Xuebiao Yao are our external reviewers.
Annotation Statistics. Reactome comprises 15,212 human reactions organized into 2,698 pathways involving 30585 proteins and modified forms of proteins encoded by 11226 different human genes, 14789 complexes, 2128 small molecules, and 1047 drugs. These annotations are supported by 38549 literature references. We have projected these reactions onto 79407 orthologous proteins, creating 19520 orthologous pathways in 14 non-human species. Version 88 has annotations for 4944 protein variants (mutated proteins) and their post-translationally modified forms, derived from 359 proteins, which have contributed to the annotation of 1802 disease-specific reactions and 725 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provide tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource as well as a Global Core Biodata Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art, and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
 Respiratory Syncytial Virus Infection Pathway
Respiratory Syncytial Virus Infection Pathway
New and Updated Topics and Pathways. As we head into winter and medical experts warn, once again, of a potential tripledemic of flu, COVID-19, and Respiratory Syncytial Virus (RSV), Reactome completes its coverage of these three viruses with the release of the RSV infection pathway in V87. Additional topics with new or revised pathways in this release include Cellular responses to stimuli (Cellular response to mitochondrial stress), Developmental Biology (Kidney formation and Specification of the neural plate border), Disease (Defects of platelet adhesion to exposed collagen, and Signaling by ALK in cancer), DNA Repair (Processing of DNA double-strand break ends), Hemostasis (Platelet Adhesion to exposed collagen), Metabolism (Sphingolipid metabolism), Metabolism of proteins (Mitochondrial protein degradation), Signal Transduction (Signaling by ALK and Signaling by EGFR), Transport of small molecules (Glycosphingolipid transport).
New and Updated Illustrations. New or revised Illustrations with embedded navigation features have been created for Cellular responses to stress, Developmental Biology, Diseases of hemostasis, Gastrulation, Kidney development, Metabolism of proteins, Transport of small molecules, Viral Infection Pathways and Respiratory Syncytial Virus Infection Pathway.
Thanks to our Contributors. Mazen Aljghami, Harrison Bergeron, Rui Gao, Lena Gunhaga, Xiaoyan Guo, David P Hill, Walid A Houry, Anthony Mak, Trevor Morey, Cedric Patthey, and Suzanne D Turner are our external reviewers.
Annotation Statistics. Reactome comprises 15,046 human reactions organized into 2,673 pathways involving 30,451 proteins and modified forms of proteins encoded by 11,180 different human genes, 14,594 complexes, 2,120 small molecules, and 1,046 drugs. These annotations are supported by 37,933 literature references. We have projected these reactions onto 79,040 orthologous proteins, creating 19,398 orthologous pathways in 14 non-human species. Version 87 has annotations for 4,942 protein variants (mutated proteins) and their post-translationally modified forms, derived from 357 proteins, which have contributed to the annotation of 1,796 disease-specific reactions and 723 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provide tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource as well as a Global Core Biodata Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

Maternal to zygotic transition
New and Updated Topics and Pathways. In V86, Topics with new or revised pathways include Cellular responses to stimuli (KEAP1-NFE2L2 pathway), Developmental biology (Formation of anterior neural plate, Formation of posterior neural plate, and Maternal to zygotic transition (MZT)), Immune system (PKR-mediated signaling), Metabolism (Vitamin B5 (pantothenate) metabolism), Metabolism of RNA (Mitochondrial RNA degradation), and Metabolism of proteins (Protein hydroxylation).
New and Updated Illustrations. New or revised Illustrations with embedded navigation features have been created for Antiviral mechanism by IFN-stimulated genes, Developmental Biology, Gastrulation, Maternal to zygotic transition, and Metabolism of RNA.
Thanks to our Contributors. David Hill, Hisato Kondoh, Rekha Patel, and Wei Xie are our external reviewers.
Annotation Statistics. Reactome comprises 14,803 human reactions organized into 2,647 pathways involving 30,338 proteins and modified forms of proteins encoded by 11,154 different human genes, 14,441 complexes, 2,025 small molecules, and 1,119 drugs. These annotations are supported by 37,156 literature references. We have projected these reactions onto 84,414 orthologous proteins, creating 19,561 orthologous pathways in 14 non-human species. Version 86 has annotations for 4,919 protein variants (mutated proteins) and their post-translationally modified forms, derived from 354 proteins, which have contributed to the annotation of 1,667 disease-specific reactions and 707 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provide tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource as well as a Global Core Biodata Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

New and Updated Topics and Pathways. In V85, Topics with new or revised pathways include Developmental Biology (Cardiogenesis, Formation of intermediate mesoderm, Formation of paraxial mesoderm, Somitogenesis, Specification of primordial germ cells ), Cell-Cell communication (Regulation of Expression and Function of Type II Classical Cadherins, Regulation of CDH11 Expression and Function, and Regulation of CDH19 Expression and Function), Cellular responses to stimuli (ATF6B (ATF6-beta) activates chaperones), Immune System (IFNG signaling activates MAPKs and Interferon gamma signaling), and Metabolism (Fructose biosynthesis and Fructose catabolism).
New and Updated Illustrations. New or revised Illustrations with embedded navigation features have been created for Developmental Biology, Gastrulation, and Reproduction.
Thanks to our Contributors. Ravindra Chalamalasetty and Terry Yamaguchi are our external authors. Julia Brasch, Ravindra Chalamalasetty, David Hill, Patrick Tam, and Terry Yamaguchi are our external reviewers.
Annotation Statistics. Reactome comprises 14,628 human reactions organized into 2,629 pathways involving 30,155 proteins and modified forms of proteins encoded by 11,396 different human genes, 14,277 complexes, 2,004 small molecules, and 1,114 drugs. These annotations are supported by 36,706 literature references. We have projected these reactions onto 84,167 orthologous proteins, creating 19,505 orthologous pathways in 14 non-human species. Version 85 has annotations for 4,919 protein variants (mutated proteins) and their post-translationally modified forms, derived from 354 proteins, which have contributed to the annotation of 1,659 disease-specific reactions and 707 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provide tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource as well as a Global Core Biodata Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

New and Updated Topics and Pathways. In V84, Topics with new or revised pathways include Developmental Biology (Formation of definitive endoderm), Gene Expression (Formation of WDR5-containing histone-modifying complexes), and Signal Transduction (Regulation of TNFR1 signaling, TNFR1-induced NF-kappa-B signaling pathway, and TNFR1-induced proapoptotic signaling). The Infectious disease pathway has been reorganized with grouping pathways for Bacterial Infection Pathways, Viral Infection Pathways, and Parasitic Infection Pathways.
New and Updated Illustrations. In parallel with the restructuring of the infectious disease pathway in V84, new or revised Illustrations with embedded navigation features have been created for Infectious disease, Bacterial Infection Pathways, Viral Infection Pathways, and Parasitic Infection Pathways. Additional disease pathways with new or updated Illustrations include Aberrant regulation of mitotic cell cycle due to RB1 defects, Defective factor IX causes hemophilia B, Defective factor VIII causes hemophilia A, Diseases of Cellular Senescence, and Diseases of Mitotic Cell Cycle. Normal pathways with new or revised illustrations include Epigenetic regulation of gene expression and Gastrulation.
Thanks to our Contributors. Toni Franjkić, Kai Ge, David P Hill, Ivana Munitic, Nazmus Salehin, Patrick Tam, and Hieu Van are our external reviewers.
Annotation Statistics. Reactome comprises 14516 human reactions organized into 2615 pathways involving 30095 proteins and modified forms of proteins encoded by 11371 different human genes, 14194 complexes, 2002 small molecules, and 1113 drugs. These annotations are supported by 36444 literature references. We have projected these reactions onto 84090 orthologous proteins, creating 19435 orthologous pathways in 14 non-human species. Version 84 has annotations for 4983 protein variants (mutated proteins) and their post-translationally modified forms, derived from 353 proteins, which have contributed to the annotation of 1,659 disease-specific reactions and 704 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provide tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource as well as a Global Core Biodata Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
Outreach Coordinator
We are looking for an experienced and enthusiastic person to lead our outreach efforts.
As Reactome Outreach Coordinator, you will lead our community outreach activities, strengthening existing collaborations and developing initiatives to foster new ones. Working with the software and curatorial teams, you will coordinate Reactome communications, including conference presentations, workshops, scientific and lay papers, and social media. You will develop education and training materials for in person and online learning and will contribute to the documentation on the Reactome web site.
Your responsibilities will include:
- Maintaining a calendar of outreach activities, including community events, workshops, appearances and other communication opportunities
- Planning and delivering training workshops on Reactome web interface, tools and application programming interfaces (APIs) and their use in large-scale data analysis. This responsibility will involve multiple public presentations per year as well as international travel
- Promotion of Reactome and communication of Reactome news through social media channels like Twitter, LinkedIn, and Facebook
- Creating and maintaining documentation and e-learning materials on our advanced tools and APIs, and their application in bioinformatic analysis
- Direct support via email and tracker of user queries on using our APIs and advanced tools
- Direct interaction with our development teams to fix bugs identified by users and find solutions to user problems
- Supporting usability testing of tools and APIs
Required Qualifications:
We are seeking a self-reliant, resourceful team player comfortable with multitasking who is open to cooperation and collaboration both within our group and in the larger bioinformatics community.
- You have a Master’s or PhD degree or equivalent experience in life sciences, preferably in bioinformatics, biochemistry, molecular biology, cancer research or a related field
- In addition to your academic training, you have at least 2 years of relevant job experience related to bioinformatics, pathway databases, analysis of biological data sets or similar. Knowledge of cancer genomics is helpful but not required
- Significant experience in teaching, training, public outreach, online help and/or event planning events is essential
- You should be comfortable speaking persuasively to large audiences as well as conducting teaching and tutorials in small group settings, both online and in person
- You have excellent English communication skills, both written and verbal, to facilitate effective communications with other team members and to communicate with external collaborators and users
Working hours can be negotiated to fit the needs of the applicant and the project. A part-time renewable contract or full-time permanent position can be considered, or a Professional Services Agreement for those candidates out of province.
To apply, please see the full posting here.
- Details
On December 15, the Global Biodata Coalition (https://globalbiodata.org/) of research funders recognized the Reactome Knowledgebase (www.reactome.org) as one of just 37 resources worldwide whose long-term funding and sustainability are critical to life science and biomedical research. See the full announcement here.
Many thanks to the selection panel at GBC for the recognition, and congratulations to the other resources who were selected.

- Details
Cytokine Signaling in Immune System

New and Updated Topics and Pathways. In V83, Topics with new or revised pathways include Developmental Biology (Formation of axial mesoderm, Formation of lateral plate mesoderm, Formation of paraxial mesoderm), Immune System (Signaling by CSF1 (M-CSF) in myeloid cells), Metabolism (Gluconeogenesis), Metabolism of proteins (Insulin processing), Metabolism of RNA (mRNA Splicing - Major Pathway), Programmed Cell Death (Regulation of necroptotic cell death), Signal Transduction (Regulation of TNFR1 signaling, TNFR1-induced NFkappaB signaling pathway, and TNFR1-induced proapoptotic signaling), and Transport of small molecules (Zinc efflux and compartmentalization by the SLC30 family). In continuing our collaborative drug annotation work with Caroline Thorn and PharmGKB, we have added the pathway Ciprofloxacin ADME under the topic of Drug Absorption, Distribution, Metabolism, and Excretion (ADME) pathways.
New and Updated Illustrations. Cytokine Signaling in Immune system, Diseases of DNA Double-Strand Break Repair, Disorders of Nervous System Development, Drug ADME, Gastrulation, GPCR downstream signalling, Loss of function of MECP2 in Rett syndrome, Pervasive developmental disorders, Platelet activation, signaling and aggregation, and SUMOylation have a new or revised Illustration with embedded navigation features.
Thanks to our Contributors. E Richard Stanley is our external author. Ravindrababu Chalamalasetty, Clément Charenton, David P Hill, Christian Mosimann, James M Murphy, Karin Prummel, Hiroshi Sasaki, E Richard Stanley, Hailin Tu, and Terry P Yamaguchi are our external reviewers.
Annotation Statistics. Reactome comprises 14,471 human reactions organized into 2,610 pathways involving 30,656 proteins and modified forms of proteins encoded by 11,442 different human genes, 14,153 complexes, 2,002 small molecules, and 1,113 drugs. These annotations are supported by 36,290 literature references. We have projected these reactions onto 83,933 orthologous proteins, creating 19,417 orthologous pathways in 14 non-human species. Version 83 has annotations for 4,977 protein variants (mutated proteins) and their post-translationally modified forms, derived from 361 proteins, which have contributed to the annotation of 1,659 disease-specific reactions and 704 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provides tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
Introducing the Reactome Research Spotlight!
In October 2022, Reactome introduced a new feature, the Reactome Research Spotlight. This monthly feature will highlight a recent standout publication that makes use of Reactome data or analysis tools in its research.
In our first spotlight, we feature the paper “Post-infusion CAR TReg cells identify patients resistant to CD19-CAR therapy” by Good et al, published in September 2022 in Nature Medicine. This paper identifies expansion of regulatory T cells as a biomarker of resistance and toxicity after CD19-CAR therapy in patients with large B cell lymphoma. Differential gene expression and Reactome pathway enrichment analysis identified TReg development as one of the top enriched pathways in the TReg CD4+ CD57-Helios+ cell populations associated with post-therapy progression and decreased neurotoxicity. Do you use Reactome data or tools in your research? Email us at This email address is being protected from spambots. You need JavaScript enabled to view it. to have your work highlighted in an upcoming Spotlight feature.
Do you use Reactome data or tools in your research? Email us at This email address is being protected from spambots. You need JavaScript enabled to view it. to have your work highlighted in an upcoming Spotlight feature.
Follow us on Twitter! @reactome to stay up to date with new and updated pathways, feature updates and more.
- Details

Reactome is pleased to announce a partnership with PharmGKB to curate pharmacokinetic / Drug ADME (absorption distribution metabolism excretion) pathways. Sharing knowledge and distributing curation effort will increase the efficiency and consistency with which both resources are able to integrate pathway data. The cross-references between co-curated pathways in Reactome and PharmGKB will support seamless navigation between the two resources enabling users to understand, visualize, and analyze their data in contexts that are best suited to their research interests. The collaborative pathways released in Reactome Version 82 are Prednisone ADME (found here at PharmGKB), and Ribavirin ADME (found here at PharmGKB).
- Details

New and Updated Topics and Pathways. In V82, Topics with new or revised pathways include Developmental Biology (Epithelial-Mesenchymal Transition during gastrulation and Germ layer formation at gastrulation), Disease-SARS-CoV-1 Infection (SARS-CoV-1-host interactions), Disease-SARS-CoV-2 Infection (Induction of Cell-Cell Fusion, Maturation of nucleoprotein, Maturation of protein E, Maturation of protein M, Maturation of Spike protein and Translation of Accessory Proteins, Gene expression (Transcriptional Regulation by NPAS4), Metabolism (HS-GAG degradation, Keratan sulfate degradation, and Transport and synthesis of PAPS), and Signal Transduction (Regulation of TNFR1 signaling and TNF signaling). In continuing our collaborative drug annotation work with Caroline Thorn and PharmGKB, we have added the pathways Prednisone ADME and Ribavirin ADME under the topic of Drug Absorption, Distribution, Metabolism, and Excretion (ADME) pathways.
New and Updated Illustrations. Drug Absorption, Distribution, Metabolism and Excretion (ADME), Gastrulation, SARS-CoV-2 infection, SUMOylation, and SUMO E3 ligases SUMOylate target proteins have a new or revised Illustration with embedded navigation features. The updated SARS-CoV-2 illustration featured above highlights the recent partitioning of the pathway into early and late subpathways based on our growing understanding of the life cycle of this virus.
Thanks to our Contributors. David Hill, Yingxi Lin, Rainer de Martin, Caroline Thorn, and Hailin Tu are our external reviewers.
Annotation Statistics. Reactome comprises 14398 human reactions organized into 2601 pathways involving 11393 proteins and modified forms of proteins encoded by 11097 different human genes, 14084 complexes, 1998 small molecules, and 1035 drugs. These annotations are supported by 35965 literature references. We have projected these reactions onto 83288 orthologous proteins, creating 19385 orthologous pathways in 15 non-human species. Version 82 has annotations for 4977 protein variants (mutated proteins) and their post-translationally modified forms, derived from 352 proteins, which have contributed to the annotation of 1659 disease-specific reactions and 704 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Services. The ReactomeFIViz app and ReactomeGSA package provide tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here..
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

New and Updated Topics and Pathways. In V81, Topics with new or revised pathways include Development (Germ layer formation at gastrulation), Immune system (TAK1-dependent IKK and NF-kB activation), and Signal Transduction (Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer). The SARS-CoV-2 life cycle has been updated to reflect our growing understanding of the life cycle. Viral events have partitioned into early and late subpathways. Early events begin with viral entry and carry through to initial gene expression; late events include the massive organelle reorganizations and viral release as host cells transition to viral shedding factories. SARS-CoV-2 annotation now includes 224 events; with this release 61 of those events have been updated with experimental support from the body of COVID-19 literature that has emerged since March 2020. Additionally, in collaboration with Rachel Huddart with PharmGKB, we have added 101 drug interactions associated with 63 pathways in this release bringing the total number of drugs represented in Reactome to 1022.
New and Updated Illustrations. Developmental Biology, Gastrulation, Nucleotide catabolism defects, and Nucleotide salvage defects have a new or revised Illustration with embedded navigation Features.
Thanks to our Contributors. Osvaldo Contreras, Rainer de Martin, Rachel Huddart, Naz Salehin, and Patrick P L Tam are our external reviewers.
Annotation Statistics. Reactome comprises 14,246 human reactions organized into 2,585 pathways involving 11,291 proteins and modified forms of proteins encoded by 11,088 different human genes, 13,984 complexes, 1,986 small molecules, and 1,022 drugs. These annotations are supported by 35,629 literature references. We have projected these reactions onto 83,266 orthologous proteins, creating 19,328 orthologous pathways in 15 non-human species. Version 81 has annotations for 5,422 protein variants (mutated proteins) and their post-translationally modified forms, derived from 352 proteins, which have been used to annotate disease-specific 1,609 reactions and 697 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeFIViz app and ReactomeGSA package provides tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The EMBL - European Bioinformatics Institute. Reactome is an ELIXIR Core Data Resource. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence applies to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question, want to provide feedback, or are interested in collaborating with us to annotate a topic, please contact us at This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

A new research article entitled “Evaluating the predictive accuracy of curated biological pathways in a public knowledgebase” has been published in the Database (Oxford) journal. The paper describes the utility of Reactome pathways for predicting functional consequences of genetic perturbations. Predictions of perturbation effects based on Reactome pathways were compared against published empirical observations. More publications from the Reactome Team can be found here.
- Details

New and Updated Topics and Pathways. In V80, Topics with new or revised pathways include Cellular responses to stimuli (Cellular response to chemical stress, Cytoprotection by HMOX1, KEAP1-NFE2L2 pathway), Disease (Defective HDR through Homologous Recombination (HRR) Due to BRCA2 Loss of Function, Diseases of nucleotide metabolism, and SARS-CoV-2-host interactions), DNA repair (Presynaptic phase of homologous DNA pairing and strand exchange), Drug ADME (Atorvastatin ADME), Metabolism (Cobalamin (Cbl, vitamin B12) transport and metabolism), and Metabolism of Proteins (Neddylation).
New and Updated Illustrations. Cellular response to chemical stress, Defects in cobalamin (B12) metabolism, Diseases of Metabolism, Diseases of Mismatch Repair (MMR), Diseases of nucleotide metabolism, Drug Absorption, Distribution, Metabolism and Excretion (ADME), Glycogen storage diseases, RAS GTPase cycle mutants, and Signaling by MRAS-complex mutants have a new or revised Illustration with embedded navigation features.
Thanks to our Contributors. Antonio Cuadrado, Wolf-Dietrich Heyer, David P Hill, Rachel Huddart, Hang Phuong Le, Jie Liu, Francesco Messina, and Julia Somers are our external reviewers.
Annotation Statistics. Reactome comprises 14,108 human reactions organized into 2,580 pathways involving 11,285 proteins and modified forms of proteins encoded by 11,084 different human genes, 13,869 complexes, 1,987 small molecules, and 532 drugs. These annotations are supported by 34,703 literature references. We have projected these reactions onto 77,392 orthologous proteins, creating 18,830 orthologous pathways in 15 non-human species. Version 80 has annotations for 5,422 protein variants (mutated proteins) and their post-translationally modified forms, derived from 352 proteins, which have been used to annotate disease-specific 1,601 reactions and 695 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeGSA package provides tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our helpdesk.
- Details

New and Updated Topics and Pathways. In V79 we have introduced the new topic Drug Absorption, Distribution, Metabolism and Excretion in which we cover both pharmacokinetics and pharmacodynamics of Aspirin, Azathioprine, and Paracetamol. Topics with new or revised pathways in this release include Cell Cycle (Cyclin D associated events in G1 and Drug-mediated inhibition of CDK4/CDK6 activity), DNA Replication (Assembly of the pre-replicative complex), Immune System (Interferon alpha/beta signaling), Metabolism (Cholesterol biosynthesis and Choline catabolism), and Sensory Perception (Expression and translocation of Olfactory Receptors).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Diseases associated with glycosaminoglycan metabolism, Diseases associated with glycosylation precursor biosynthesis,Diseases associated with N-glycosylation of proteins, Diseases of Telomere Maintenance, DNA Replication, Drug Absorption, Distribution, Metabolism, and Excretion (ADME), Drug resistance of ALK mutants, Drug resistance of FLT3 mutants, Drug resistance of KIT mutants, Drug resistance of PDGFR mutants, Metabolism, Signaling by AMER1 mutants, Signaling by APC mutants, Signaling by AXIN mutants, Signaling by CTNNB1 phospho-site mutants.
Thanks to our Contributors. Our external authors are Jelena Kusic-Tisma and Damjana Rozman. Our external reviewers are Andrew J Brown, Rachel Huddart, Anne Morgat, Sisira Kadambat Nair, Alexander F Palazzo, Francesco Raimondi, Qingtang Shen.
Annotation Statistics. Reactome comprises 13,960 human reactions organized into 2,553 pathways involving 11,270 proteins and modified forms of proteins encoded by 11,071 different human genes, 13,853 complexes, 1,987 small molecules, and 527 drugs. These annotations are supported by 34,252 literature references. We have projected these reactions onto 77,418 orthologous proteins, creating 19,597 orthologous pathways in 15 non-human species. Version 79 has annotations for 5,064 protein variants (mutated proteins) and their post-translationally modified forms, derived from 347 proteins, which have been used to annotate disease-specific 1,540 reactions and 674 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeGSA package provides tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our helpdesk.
- Details
 Our user community is built on success stories! There are thousands of you around the world and many of you have created amazing experiments, tools and resources using Reactome. We're always looking for ways to share interesting discoveries made with Reactome, and have decided to showcase projects on our website to inspire others to create new stories. If you have a novel success story or use case you'd like to share, we would love to hear from you.
Our user community is built on success stories! There are thousands of you around the world and many of you have created amazing experiments, tools and resources using Reactome. We're always looking for ways to share interesting discoveries made with Reactome, and have decided to showcase projects on our website to inspire others to create new stories. If you have a novel success story or use case you'd like to share, we would love to hear from you.
To share your use case or success story, please fill in the success story form or email us at This email address is being protected from spambots. You need JavaScript enabled to view it..
What is required?
There is no specific experiment, project or resource that we are looking for. We are mostly interested in hearing from you if you are willing to share what you have learned or what has worked well.
Some potential stories............
- describe how Reactome data or software was integrated into your data resource or software tool?
- tell of how using Reactome helped you or your group to make a valuable discovery about your experiment?
Where will my story be shared?
Your story will be reviewed by members of the Reactome team and may be shared on reactome.org. This will essentially be a post written by you and one of our team members. Your story will also be shared on Twitter.
What will be the process?
Filling out this form will simply indicate your interest in being featured on Reactome. If your submission is selected, we'll then email you for more detailed information about your success story and experiences. We will publish a novel success story every month. Every year, one entry from the "Success Story of the Month" will be selected to receive one $100 gift card redeemable through Amazon or another online vendor.
- Details

New and Updated Topics and Pathways. Topics with new or revised pathways in V78 include Disease (Defective HDR through homologous recombination (HRR) due to BRCA1 loss of function and Defective HDR through homologous recombination (HRR) due to PALB2 loss of function), DNA repair (Alkylating DNA damage induced by chemotherapeutic drugs, Drug-induced formation of DNA interstrand crosslinks, and Reversible DNA damage induced by alkylating chemotherapeutic drugs), DNA Replication (Assembly of the ORC complex at the origin of replication), Sensory Perception (Olfactory signaling and Sensory perception of taste), and Signal transduction (CDC42 GTPase Cycle, Drug-mediated inhibition of MET activation, Met Receptor Activation, RHO GTPases regulate CFTR trafficking, RHOJ GTPase cycle, RHOQ GTPase cycle, and TGF-beta receptor signaling activates SMADs).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Defective intrinsic pathway to apoptosis, Defects in biotin (Btn) metabolism, Defects in cobalamin (B12) metabolism, Defects of contact activation system (CAS) and kallikrein/kinin system (KKS), Diseases associated with O-glycosylation of proteins, Diseases associated with N-glycosylation of proteins, Diseases associated with the TLR signaling cascade, Diseases of base excision repair, Diseases of programmed cell death, DNA replication, Loss of function of SMAD2/3 in cancer, Mucopolysaccharidoses, Pentose phosphate pathway disease, Sensory perception, Signaling by NOTCH1 in cancer, and Signaling by Rho GTPases, Miro GTPases and RHOBTB3.
Thanks to our Contributors. Our external authors are Jelena Kusic-Tisma, Sisira Kadambat Nair. Our external reviewers are Kaja Blagotinšek Cokan, Maitreyi E Das, Peihua Jiang, Jean-Yves Masson, Larissa Milano, Gemma Montalban, Sisira Kadambat Nair, and Francesco Raimondi.
Annotation Statistics. Reactome comprises 13,890 human reactions organized into 2,546 pathways involving 10,918 proteins and modified forms of proteins encoded by 10,720 different human genes, 13,804 complexes, 1,940 small molecules, and 507 drugs. These annotations are supported by 34,025 literature references. We have projected these reactions onto 77,335 orthologous proteins, creating 18,698 orthologous pathways in 15 non-human species. Version 78 has annotations for 4,603 protein variants (mutated proteins) and their post-translationally modified forms, derived from 352 proteins, which have been used to annotate disease-specific 1,544 reactions and 673 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and access programmatically through our Content and Analysis Services. The ReactomeGSA package provides tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our helpdesk.
- Details
 On June 2, 2021, Dr. Johannes Griss presented "A guide to multiomics pathway analysis" to a record audience of 572 participants as part of the EMBL-EBI webinar series. The seminar recording and training materials, which are now available online, provides insight into performing comparative multi-omics pathway analyses using the ReactomeGSA platform. We want to take this opportunity to thank all those that took part in the training webinar. If you are interested in learning more about Reactome or if you have any additional questions, please This email address is being protected from spambots. You need JavaScript enabled to view it..
On June 2, 2021, Dr. Johannes Griss presented "A guide to multiomics pathway analysis" to a record audience of 572 participants as part of the EMBL-EBI webinar series. The seminar recording and training materials, which are now available online, provides insight into performing comparative multi-omics pathway analyses using the ReactomeGSA platform. We want to take this opportunity to thank all those that took part in the training webinar. If you are interested in learning more about Reactome or if you have any additional questions, please This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

New and Updated Topics and Pathways. Topics with new or revised pathways in V77 include Disease (Defective HDR through Homologous Recombination (HRR) Due to PALB2 Loss of Function, Defective pyroptosis, Loss of Function of TP53 in Cancer, Maturation of nucleoprotein, and Signaling by ALK in cancer), DNA repair (Homologous DNA Pairing and Strand Exchange), Programmed Cell Death (Pyroptosis), and Signal transduction (Fc epsilon receptor (FCERI) signaling, RHOD GTPase Cycle, RHOF GTPase Cycle, RHOU GTPase Cycle, RHOV GTPase Cycle, and Signaling by ALK).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Cellular response to chemical stress, Cellular response to starvation, Cellular responses to stress, Diseases of glycosylation, Diseases of Signal Transduction by growth factor receptors and second messengers, HIV Infection, Metabolism of proteins, Oncogenic MAPK signaling, Regulated Necrosis, Signaling by ALK in Cancer, Signaling by FGFR in Disease, Signaling by KIT in Disease, Signaling by receptor tyrosine kinases, and Signaling by Rho GTPases.
Thanks to our Contributors. Our external reviewers are Marcio Luis Acencio, Mariano Bisbal, Giorgio Inghirami, Anna Niarakis, Helmut Pospiech, Kazuyasu Sakaguchi, Mikhail V Shepelev, Feng Shao, and Robert Winqvist.
Annotation Statistics. Reactome comprises 13,827 human reactions organized into 2,536 pathways involving 11,374 proteins and modified forms of proteins encoded by 11,084 different human genes, 13,662 complexes, 1,857 small molecules, and 432 drugs. These annotations are supported by 33,752 literature references. We have projected these reactions onto 78,539 orthologous proteins, creating 18,659 orthologous pathways in 15 non-human species. Version 77 has annotations for 4,464 protein variants (mutated proteins) and their post-translationally modified forms, derived from 347 proteins, which have been used to annotate disease-specific reactions and pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeGSA package provides tools for multi-omics data analysis. The idg.reactome.org Web Portal provides a collection of web-based tools to help researchers place understudied proteins in a pathway context.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our helpdesk.
- Details

As part of the Cutting Edge Informatics Tools (CEIT) for the Illuminating the Druggable (IDG) Program, the Reactome group has built a new tool, the Reactome IDG Portal, that utilizes the Reactome knowledgebase to systematically illuminate interactions of dark proteins with other proteins and biological entities, allowing evaluation of these understudied proteins via their localizations and potential interactions, and facilitating the design of experiments to test their functions. The portal enables users to:
- Search any gene and view its location in Reactome’s pathways based on manual annotation or interactions via one-hop pairwise relationships.
- Convert biochemical reaction-based diagrams into simple pairwise networks.
- View scored interacting pathways based on functional interactions predicted from a random forest model trained with 106 features.
- Construct new overlays and visualizations for protein-protein pairwise relationships or drug-target interactions.
- Use an extended diagram viewer to visualize protein knowledge levels, and overlay multiple tissue-specific expression values from 19 data sources from Target Central Resource Database (TCRD).
Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, New York University Langone Medical Center, Oregon Health and Science University, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website. Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more.
The Illuminating the Druggable Genome (IDG) Program, launched by the National Institutes of Health (NIH) Common Fund, is a multidisciplinary project to improve the scientific understanding of understudied members of three key protein families: non-olfactory G-protein-coupled receptors (GPCRs), ion channels and protein kinases. The overall goal of the IDG Program is to catalyze research in areas of biology that are currently understudied but that have high potential to impact human health. Follow us on Twitter: @DruggableGenome to receive updates about the program.
- Details

New and Updated Topics and Pathways. Topics with new or revised pathways in V76 include Cellular responses to external stimuli (Cytoprotection by HMOX1 and Heme signaling), Disease (Defective pyroptosis), Gene Expression (tRNA-derived small RNA (tsRNA) biogenesis), Immune system (DDX58/IFIH1-mediated induction of interferon-alpha/beta and Signaling by CSF3 (G-CSF)), Programmed cell death (Pyroptosis), Sensory Perception (Sensory processing of sound), and Signal transduction (Miro GTPase Cycle, RHO GTPase Cycle, and RHOH GTPase Cycle).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Cellular responses to stress, Cytokine Signaling in Immune system, Diseases of Metabolism, Metabolic disorders of biological oxidations enzymes, Sensory Perception, and Sensory processing of sound.
Thanks to our Contributors. Our external reviewers are Katia Basso, Ines Castro, Peter Dallos, Anindya Dutta, Philippe Fort, David N. Furness, Thirumala-Devi Kanneganti, Carlos Henrique Inacio Ramos, Michael Schrader, Hans-Uwe Simon, Julia Somers, Zhangli Su, Ivo P. Touw, Briana Wilson, and Zhibin Zhang.
Annotation Statistics. Reactome comprises 13,732 human reactions organized into 2,516 pathways involving 11,362 proteins and modified forms of proteins encoded by 11,073 different human genes, 13,409 complexes, 1,856 small molecules, and 415 drugs. These annotations are supported by 33,453 literature references. We have projected these reactions onto 74,384 orthologous proteins, creating 18,051 orthologous pathways in 15 non-human species. Version 76 has annotations for 2,949 protein variants (mutated proteins) and their post-translationally modified forms, derived from 337 proteins, which have been used to annotate disease-specific 1,550 reactions and 653 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape and the ReactomeGSA package provides tools for multi-omics data analysis.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our helpdesk.
- Details

New and Updated Pathways. Topics with new or revised pathways in release 75 include Developmental biology (Transcriptional regulation of testis differentiation), Disease (Alternative lengthening of telomeres, Defective DNA double strand break response due to BARD1 loss of function, Defective DNA double strand break response due to BRCA1 loss of function, Signaling by FLT3 in disease), DNA Repair (Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks), Immune system (FLT3 signaling), and Signal transduction (RHOBTB GTPase cycle).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Developmental biology, Diseases associated with visual transduction, Diseases of DNA repair, Diseases of hemostasis, Diseases of the immune system, Diseases of mitotic cell cycle, Diseases of signal transduction by growth factor receptors and second messengers, FLT3 signaling in disease, Hh mutants abrogate ligand secretion, PI3PK/AKT signaling in cancer, Selective autophagy, Signaling by ERBB2 in cancer, Signaling by PDGFR in disease, Signaling by WNT in cancer, and Uptake and actions of bacterial toxins.
Thanks to our Contributors. Our external authors are Richard J Baer and Francisco Rivero Crespo. Our external reviewers are Richard J Baer, Kenya Imaimatsu, Yoshiakira Kanai, Julhash Kazi, Alan Meeker, Roger Reddel, and Francisco Rivero Crespo.
Annotation Statistics. Reactome comprises 13,534 human reactions organized into 2,477 pathways involving 11,118 proteins and modified forms of proteins encoded by 10,929 different human genes, 13,210 complexes, 1,854 small molecules, and 414 drugs. These annotations are supported by 32,493 literature references. We have projected these reactions onto 79,333 orthologous proteins, creating 18,510 orthologous pathways in 15 non-human species. Version 75 has annotations for 2,620 protein variants (mutated proteins) and their post-translationally modified forms, derived from 327 proteins, which have been used to annotate disease-specific 1,297 reactions and 605 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our helpdesk.
- Details

A new research article entitled “ReactomeGSA - Efficient Multi-Omics Comparative Pathway Analysis” has been published in the Molecular & Cellular Proteomics journal. The paper discusses the novel ReactomeGSA resource for comparative pathway analyses of multi-omics datasets using our existing web interface and a novel R Bioconductor package with explicit support for scRNA-seq data. The ReactomeGSA tools are described here. More publications from the Reactome Team can be found here.
- Details

New Feature. In response to the COVID-19 pandemic, Reactome is fast-tracking the annotation of Human Coronavirus infection pathways in collaboration with the COVID-19 Disease Map group. Reactome release 74 features the SARS-CoV-2 (COVID-19) infection pathway. To generate this pathway, we started with a rough draft generated computationally via our orthoinference process from the previously manually curated, peer-reviewed Reactome SARS-CoV-1 (Human SARS coronavirus) infection pathway. The draft SARS-CoV-2 (COVID-19) infection pathway events and entities were then reviewed by Reactome curators and curated using published SARS-CoV-2 experimental data. Before finalization, the resulting curated SARS-CoV-2 infection pathway was peer-reviewed by external experts. The current pathway consists of 101 reactions involving 489 molecular entities (279 proteins, 12 RNAs and 198 other), and is supported by citations to 227 publications. In future releases of Reactome, we will extend our annotation of the pathway, indicate drugs and other compounds that modulate steps in infection, and add molecular events that link infectious pathway steps to host immune processes and other aspects of human biology that determine responses to viral infection. This accelerated annotation project is supported by a recently-received supplement grant U41 HG003751-13S1 from the National Human Genome Research Institute.
New and Updated Pathways. Other topics with new or revised pathways in release 74 include Disease (Defective RIPK1-mediated regulated necrosis) and Programmed cell death (RIPK1-mediated regulated necrosis).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Diseases of metabolism, SARS-CoV infections, ABC transporter disorders, Diseases associated with surfactant metabolism, Diseases of cellular response to stress, Diseases of DNA repair, Diseases of mitotic cell cycle, Diseases of neuronal system, Diseases of signal transduction by growth factor receptors and second messengers, Disorders of developmental biology, Disorders of transmembrane transporters, Signaling by TGF-beta receptor complex in cancer, and SLC transporter disorders. New static illustrations are now available for Complement cascade, Rho GTPase cycle, SARS-CoV-2 infection, and Toll-Like receptors cascades.
Thanks to our Contributors. Our external author is Andrea Senff-Ribeiro and our external reviewers are Marcio Luis Acencio, Najoua Lalaoui, James M Murphy.
Annotation Statistics. Reactome comprises 13,416 human reactions organized into 2,441 pathways involving 11,110 proteins and modified forms of proteins encoded by 10,922 different human genes, 12,976 complexes, 1,854 small molecules, and 428 drugs. These annotations are supported by 32,297 literature references. We have projected these reactions onto 79,190 orthologous proteins, creating 18,462 orthologous pathways in 15 non-human species. Version 74 has annotations for 2,624 protein variants (mutated proteins) and their post-translationally modified forms, derived from 328 proteins, which have been used to annotate disease-specificreactions and pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

New Feature. In response to the COVID-19 pandemic, Reactome is fast-tracking the annotation of Human Coronavirus infection pathways in collaboration with the COVID-19 Disease Map group. V73 includes descriptions of the SARS-CoV-1 Infection pathway as well as Potential therapeutics for SARS. In the coming months, we will use these descriptions as starting points to annotate the SARS-CoV-2 infection pathway and interactions with host proteins that affect the severity of COVID-19 disease.
New and Updated Pathways. Other topics with new or revised pathways in V73 include Cell Cycle (Inhibition of DNA recombination at telomeres, Telomere Maintenance), Disease (Aberrant regulation of mitotic cell cycle due to RB1 defects, Defects of contact activation system (CAS) and kallikrein/kinin system (KKS), Oncogenic MAPK signaling, Signaling by KIT in disease, and Signal Transduction (RAF/MAP kinase cascade, RAS processing).
New and Updated Illustrations. Illustrations with embedded navigation features have been added for Nervous system development, Defects in vitamin and cofactor metabolism, Diseases of Carbohydrate metabolism, Diseases of Metabolism, and SARS-CoV infections. New static illustrations are now available for Costimulation by the CD28 family, CD28 co-stimulation, Downstream TCR signaling, G2/M DNA damage checkpoint, G2/M DNA replication checkpoint, Generation of second messenger molecules, p53-Independent DNA Damage Response, Phosphorylation of CD3 and TCR zeta chains, SARS-CoV-1 Infection, and Translocation of ZAP-70 to Immunological synapse.
Thanks to our Contributors. Marcio Luis Acencio, Frederick A Dick, Alfonso García-Valverde, Evripidis Gavathiotis, Makoto T Hayashi, Alexander Mazein, Daniel Pilco-Janeta, Cesar Serrano, Brian Shoichet, and Bin Zhang are our external reviewers.
Annotation Statistics. Reactome comprises 13,248 human reactions organized into 2,423 pathways involving 11,111 proteins and modified forms of proteins encoded by 10,923 different human genes, 12,728 complexes, 1,869 small molecules, and 369 drugs. These annotations are supported by 32,150 literature references. We have projected these reactions onto 81,835 orthologous proteins, creating 18,654 orthologous pathways in 15 non-human species. Version 73 has annotations for 2,620 protein variants (mutated proteins) and their post-translationally modified forms, derived from 327 proteins, which have been used to annotate disease-specific 1,297 reactions and 605 pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
 A new research article entitled “Using Reactome to build an autophagy mechanism knowledgebase” has been published in the Autophagy journal. The paper discusses the curation and annotation of the molecular mechanisms of autophagy. The autophagy pathway can be viewed here. More publications from the Reactome Team can be found here.
A new research article entitled “Using Reactome to build an autophagy mechanism knowledgebase” has been published in the Autophagy journal. The paper discusses the curation and annotation of the molecular mechanisms of autophagy. The autophagy pathway can be viewed here. More publications from the Reactome Team can be found here.
- Details

Data science represents a new and evolving way of doing scientific research with critical elements including reproducibility, open access to data, methods and source code, and highly reusable and modular services on the web.
Given the popularity of the Python programming language among data scientists, we have created the reactome2py package, which facilitates communication between our tools and web services with Python. The reactome2py package consists of a library of helper functions that wrap calls to the Reactome RESTful API and a utility module that simplifies access to our open-data.
The Pathway Analysis Service pathway over-representation and expression analysis as well as a species comparison tool. Further details regarding the AnalysisService API calls are available here. The Content Service provides access to our data via an easy API based on the Representational State Transfer (REST) protocol. Finally, Utility (Utils) provides functions for fetching pathway, drug, and drug-target data, other annotations, mapping information, and overlay data from human networks. Further details regarding the Data model key classes are available here. Explore our tools and web services and learn how to include them in your applications at our Developer's Zone.
- Details

A new research article entitled “Science Forum: Wikidata as a knowledge graph for the life sciences” has been published in the eLife journal. The paper describes the use of Wikidata as a platform for the integration of biological knowledge. Over 2,200 pathways from Reactome were integrated into the Wikidata repository. More publications from the Reactome Team can be found here.
- Details

New and Updated Pathways. In version 72, topics with new or revised pathways include Autophagy (Pexophagy), Cell Cycle (Nuclear Envelope Reassembly), Developmental Biology (EGR2- and SOX10-mediated initiation of Schwann cell myelination), Disease (Signaling by ERBB2 in Cancer, Signaling by ERBB2 ECD Mutants, Signaling by ERBB2 TMD/JMD Mutants, Signaling by PDGFR in disease, and Leishmania infection), and Signal Transduction (NGF-stimulated transcription and Drug-mediated inhibition of ERBB2 signaling).
New and Updated Illustrations. Illustrations with embedded navigation features have been added or revised for Disease, Infectious disease, and Leishmania infection and Developmental Biology. New static illustrations are now available for Regulation of commissural axon pathfinding by SLIT and ROBO, Signaling by ROBO receptors, and TCR signaling.
Thanks to our Contributors. John Aletta, Ron Bose, Larry Gerace, David Gregory, Carman Ip, Rama Krishna Kancha, Therese Kinsella, Anagha Krishna, Xiangguo Liu, Anne Martin, Emmanouil Metzakopian, Adam Miller, Eamon Mulvaney, Kirk Staschke, and Shun Yao are our external reviewers.
Annotation Statistics. Reactome comprises 12,986 human reactions organized into 2,362 pathways involving 11,096 proteins and modified forms of proteins encoded by 10,908 different human genes, 12,728 complexes, 1,865 small molecules, and 237 drugs. These annotations are supported by 31,237 literature references. We have projected these reactions onto 83,698 orthologous proteins, creating 18,996 orthologous pathways in 15 non-human species. Version 72 has annotations for 1,890 protein variants (mutated proteins) and their post-translationally modified forms, derived from 315 proteins, which have been used to annotate disease-specific reactions and pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

A new Perspective/Opinion article titled “Reactome and ORCID-fine-grained credit attribution for community curation” has been published in the Database (Oxford) journal. The paper describes how we are using ORCID identifiers to provide clear credit attribution for authors, curators and reviewers to support community engagement. More publications from the Reactome Team can be found here.
- Details

New and Updated Pathways. In version 71, Topics with new or revised pathways include Autophagy (Aggrephagy), Cell Cycle (EML4 and NUDC in mitotic spindle formation), Cellular responses to external stimuli (EIK2AK4 (GCN2) modifies gene expression in response to amino acid deficiency and Response of EIF2AK1 (HRI) to heme deficiency), Disease (Defective base excision repair associated with OGG1, HCMV infection, Infection with Mycobacterium tuberculosis, and Signaling by ERBB2 in Cancer), Generic transcription pathway (Transcriptional regulation by VENTX), Metabolism (Metabolism of porphyrins), and Signal Transduction (Drug-mediated inhibition of ERBB2 signaling).
New and Updated Illustrations. Biosynthesis of specialized proresolving mediators (SPMs), Fatty acid metabolism, Metabolism, Metabolism of lipids, Metabolism of vitamins and cofactors, and Phospholipid metabolism have new Illustrations with embedded navigation features. Cellular responses to stress, Digestion and absorption, and Autophagy have revised Illustrations with embedded navigation features.
Thanks to our Contributors. Ralf Stephan is our external author. Susanne Bechstedt, Istvan Boldogh, Alain Bruhat, Patrizia Caposio, Jane-Jane Chen, Armin Deffur, Andrew Fry, Rama Krishna Kancha, Kellie Lucken, Emmanouil Metzakopian, Laura O'Regan, Daniel Streblow, Naidu M Vegi, Spiros Vlahopoulos, and Robert Wilkinson are our external reviewers.
Annotation Statistics. Reactome comprises 12,608 human reactions organized into 2,282 pathways involving 11,053 proteins and modified forms of proteins encoded by 10,870 different human genes, 12,489 complexes, 1,863 small molecules, and 225 drugs. These annotations are supported by 30,721 literature references. We have projected these reactions onto 83,392 orthologous proteins, creating 18,697 orthologous pathways in 15 non-human species. Version 71 has annotations for 1,816 protein variants (mutated proteins) and their post-translationally modified forms, derived from 310 proteins, which have been used to annotate disease-specific reactions and pathways.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

A new research article titled “The reactome pathway knowledgebase” has been published in the forthcoming 2020 NAR Database Issue. The paper describes annotating the molecular mechanisms of drug action, facilitating community involvement in annotation, and improvements to the reaction and pathway visualization. More publications from the Reactome Team can be found here.
- Details

New and Updated Pathways. In version 70, topics with new or revised pathways include Cellular response to stress (Amino acids regulate mTORC1), Disease (Evasion of oncogene induced senescence due to p16-INK4A defects, Evasion of oxidative stress induced senescence due to p16-INK4A defects, Defective intrinsic pathway for apoptosis due to p14ARF loss of function, Evasion of oncogene induced senescence due to p14ARF defects, Evasion of oxidative stress induced senescence due to p14ARF defects), Immune System (Alpha-protein kinase 1 signaling pathway and FLT3 signaling), and Signal transduction (NR1H2 and NR1H3-mediated signaling).
New and Updated Illustrations. Topics with new or revised illustrations include Metabolism of amino acids and derivatives and Innate Immune System.
Thanks to our Contributors. Dorothy Bennett, Kendall Condon, Carolyn Cummins, Nicholas Hayward, Sunil Joshi, Vaishnavi Nathan, Joyce Repa, Helen Rizos, David Sabatini, and Feng Shao are our external reviewers.
Annotation Statistics. Reactome comprises 12,608 human reactions organized into 2,282 pathways involving 11,040 proteins and modified forms of proteins encoded by 10,860 different human genes, 12,335 complexes, 1,856 small molecules, and 222 drugs. These annotations are supported by 30.398 literature references. We have projected these reactions onto 83,183 orthologous proteins, creating 18,679 orthologous pathways in 15 non-human species. Version 70 has annotations for 1,762 protein variants (mutated proteins) and their post-translationally modified forms, derived from 308 proteins. These have been used to annotate 536 complexes and 970 disease-specific reactions organized into 484 pathways and subpathways, and tagged with 387 Disease Ontology terms.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
As part of our continuous effort to provide visually attractive and more user-friendly access to our biological pathways, we are pleased to announce our new high level pathways overview visualisation based on Voronoi tessellation.
Following any pathway analysis, the pathway overview provides a new icon![]() which leads to a comprehensive, highly visual, interactive overview of pathway analysis results. The functionality is available on both desktop and mobile platforms.
which leads to a comprehensive, highly visual, interactive overview of pathway analysis results. The functionality is available on both desktop and mobile platforms.
- Details

Reactome is a curated database, it critically depends on the expertise of curators and domain experts as volunteer pathway authors and reviewers. Over 700 scientists have so far contributed to Reactome content. If you are one of them, we have great news for you: Your Reactome contribution can be included directly into your ORCID profile. ORCID provides a persistent digital identifier that distinguishes you from every other researcher and, through integration in key research workflows such as manuscript and grant submission, supports automated linkages between you and your professional activities ensuring that your work is recognized.
Because we appreciate your work contribution either authoring or reviewing a pathway or reaction, we have now made it easy to claim your contribution in ORCID. Using this new feature that has been integrated into our website, contributors who have provided their ORCID ID can, by simply clicking a button, reference their work in their profile.
Follow this link to learn more about how you can claim your work.
- Details

New and Updated Pathways. In version V69, Autophagy, including Chaperone Mediated Autophagy and Microautophagy is new. Topics with new or revised pathways include Development (Transcriptional Regulation of Granulopoiesis), Disease (Evasion of Oncogene Induced Senescence Due to p16-INK4A Defects and Evasion of Oxidative Stress Induced Senescence Due to p16-INK4A Defects), Immune system (FLT3 Signaling), and Signal Transduction (Extra-Nuclear Estrogen Signaling and Signaling by ERBB4).
New Illustrations. Illustration with embedded navigation features is now available for Autophagy.
Thanks to our Contributors. David Stern is our external author. Filippo Acconcia, Dorothy Bennett, Maria Marino, Emmanouil Metzakopian, Julia Skokowa, and Elie Traer are our external reviewers.
Annotation Statistics. Reactome comprises 12,505 human reactions organized into 2,272 pathways involving 11,009 proteins and modified forms of proteins encoded by 10,833 different human genes, 1,857 small molecules, and 196 drugs. These annotations are supported by 30.027 literature references. We have projected these reactions onto 81,631 orthologous proteins, creating 18,610 orthologous pathways in 15 non-human species. Version 69 has annotations for 1,612 protein variants (mutated proteins) and their post-translationally modified forms, derived from 305 proteins. These have been used to annotate 532 complexes and 968 disease-specific reactions organized into 478 pathways and subpathways, and tagged with 374 Disease Ontology terms.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
“Fostering open source collaboration with technical writers” – Season of Docs
Established upon the reputation of open source community programs like Google Summer of Code and Google Code-in, Google is introducing a new initiative called Season of Docs. The Reactome project has applied to participate in this year's Season of Docs.
Season of Docs aims to provide open source projects with an opportunity to engage with the technical writing community and for technical writers an occasion to gain experience in contributing to open source projects.
Together, we will raise community awareness of open documentation, technical writing, and how we can collectively work together to improve open source projects. Reactome looks forward to the opportunity that Season of Docs presents and for the opportunity to bring our entire community closer together.
Project Information
Project Name: Reactome
Project Description: Reactome is a free, open-source, curated and peer-reviewed pathway database. Our goal is to provide intuitive bioinformatics tools for the visualization, interpretation, and analysis of pathway knowledge to support basic research, genome analysis, modelling, systems biology, and education.
Project Website: https://reactome.org
Project Idea #1 Name: Revising the Reactome User Guide
Description: The Reactome User Guide provides an introduction to Reactome, the user interfaces and the database content. Exercises are also provided to help user practice what they have learned. New functionality and improvements to the core website and software are constantly added to the User Guide. The outcome of this project is the revision of the existing User Guide, with updated and new tutorials,a set of how-to guides to support navigating the website and using the pathway visualization and analysis tools
Docs Link: https://reactome.org/userguide
Contacts: Robin Haw: This email address is being protected from spambots. You need JavaScript enabled to view it. & Marc Gillepsie: This email address is being protected from spambots. You need JavaScript enabled to view it.
Project Idea #2 Name: Updating the Reactome Curator Guide
Description: The Reactome Curator Guide contains an overview of the curatorial process as well as a step by step guide for annotating Reactome pathways. The curator guide takes new curators through the process of project design, entry, review, and release. Each one of these steps requires that the curator is familiar with the Reactome data model, software entry tools, process pipeline, and project management. In this sense, the curator guide serves as a compendium of learned lessons, curator philosophy, and a step-by-step process blueprint. The outcome of this project is the complete revision of the current guide to reflect new data entry tools, update the current glossary of terms to accommodate changes to data model, and create short tutorials that can be used to reinforce each learning step.
Contacts: Marc Gillepsie This email address is being protected from spambots. You need JavaScript enabled to view it. and Lisa Matthews This email address is being protected from spambots. You need JavaScript enabled to view it.
More Information
For more information about Reactome and Season of Docs, please refer to the following pages:
Reactome Documentation - Learn more about Reactome docs.
Season of Docs - What is Season of Docs all about?
Guides - How it all comes together.
Timeline - Please be aware of the schedule!
Rules - The ever important rulebook.
- Details

The Reactome project provides a suite of tools for our users to perform pathway- and network-based data analysis via its web site and ReactomeFIViz, the functional interaction (FI) network-based Cytoscape app.
The Reactome FI network is built upon curated pathways in Reactome, supplemented with curated pathways from other pathway databases such as KEGG PATHWAY and PANTHER pathways. These curated pathway interactions are then integrated with gene-gene interactions derived from curated and high-throughput sources using machine learning to generate a reliable set of high-probability functional interactions.
Recently, we have released the 2018 version of the Reactome FI network. To construct this FI network, we improved the training of the Naive Bayesian Classifier, which resulted in an increase in the recall rate. Consequently, the SwissProt coverage was increased from 12,441 to 13,469 proteins (8.3% increase) and the total number of functional interactions among genes increased from 241,338 to 262,321 (8.7% increase). We attributed this to an increase in the number of predicted FIs due to the improved recall rate.
In addition, the ReactomeFIViz app (version 7.2.0) now supports the 2018 version of the FI network and contains updated Reactome Pathways dataset (Release 67). The new ReactomeFIViz app is available to download within the Cytoscape software, or directly from the Cytoscape app Store. The User Guide provides instructions on using the Reactome FI network and ReactomeFIViz app.
- Details

We are hosting a webinar aimed at new Reactome users who would like to get an idea of our data and tools and at existing Reactome users who would like to learn about the latest Reactome updates. Reactome is a freely available curated database of human biological pathways and reactions, which is updated on a regular basis. Release 68 features new and updated pathway and reaction annotations for 16 species, as well as a suite of tools for data analysis and visualization.
Event Date: 5th April 2019.
Event Time: 11.00 a.m. to 12.00 p.m. EDT.
Event Contact: Robin Haw at This email address is being protected from spambots. You need JavaScript enabled to view it..
Requisites: This webinar will be hosted using Cisco WebEx. You will need a computer, an Internet connection and a telephone/Skype connection/microphone to join an online session.
Please register at https://forms.gle/QzeG85hcfrX34ivUA
When you register, please make sure to provide your name and valid email address so that we can send you an email with the webinar details and instructions.
Feel free to pass this invitation along to colleagues who may benefit from learning about this valuable resource.
- Details

Biological processes are remarkably well-conserved over large evolutionary distances. At a practical level, this fact enables the extrapolation of mechanistic insights from species to species. The Reactome project aims to annotate the molecular details of a broad range of human biological processes based on experimental data from human systems. To link these human annotations at a molecular level to their conserved counterparts in model organism systems we use protein sequence orthology relationships to ask, for each human reaction and each model organism, whether the human proteins involved in the reaction have orthologs in the model organism. If the orthologs exist, we computationally infer the corresponding reaction for the model organism and in this way build up a predicted pathway knowledgebase for the organism. If the human protein functions as part of a complex, we search for orthologs of all components of the complex, and computationally infer the existence of the complex in the model organism if model organism counterparts of at least 75% of the human proteins are found.
With our March, 2019 (version 68) release, we have made two changes to improve the quality and usability of these inferred pathways.
First, with the development of Plant Reactome, a substantial body of rice (Oryza sativa) pathways annotated from plant experimental evidence is now available online. That material improves on orthology-based inferences from human data, and provides a better starting point for making such inferences to other plant species, so all inferences for plant species will now be generated, maintained, and made available through Plant Reactome.
Second, we are now using the PANTHER resource (Protein Analysis Through Evolutionary Relationships) to identify model organism orthologs of human proteins annotated in Reactome. This change will allow us to exploit features of PANTHER, such as the identification of least-diverged orthologs in model organism protein families, to improve the specificity of our inferences. The change is also part of a larger project to better align the Reactome with the Gene Ontology, PANTHER, the Alliance of Genome Resources, and related resources to generate interactive, expert-curated, actively maintained, and tightly integrated community genomics resources.
- Details

New and Updated Pathways. In version V68, topics with new or revised pathways include Disease (Defective Base Excision Repair Associated with NTHL1, Defective Base Excision Repair Associated with NEIL1, and Defective Base Excision Repair Associated with NEIL3), Metabolism (Blood group systems biosynthesis), Neuronal System (Assembly and cell surface presentation of NMDA receptors), Protein localization (Class I peroxisomal membrane protein import and Insertion of tail-anchored proteins into the endoplasmic reticulum membrane), and Signal Transduction (Non-genomic estrogen signaling).
New Illustrations. Illustrations with embedded navigation features are now available for Signaling by Non-Receptor Tyrosine Kinases, Signaling by Nuclear Receptors, and SUMO E3 ligases SUMOylate target proteins.
Thanks to our Contributors. Subhrajit Bhattacharya, Chad R Camp, Richarda de Voer, Evelina DeLaurentis, Paul W Doetsch, Ákos Farkas, Marc Fransen, Roland Kuiper, Ellis R Levin, Taei Matsui, Huaiyu Mi, Barbara Rivera, Harini Sampath, Blanche Schwappach, Ralf Stephan, Stephen F Traynelis, and Jia Zhou are our external reviewers.
Annotation Statistics. Reactome comprises 12,416 human reactions organized into 2,255 pathways involving 11,000 proteins and modified forms of proteins encoded by 10,825 different human genes, 1,854 small molecules, and 202 drugs. These annotations are supported by 29,885 literature references. We have projected these reactions onto 81,875 orthologous proteins, creating 18,505 orthologous pathways in 15 non-human species. Version 68 has annotations for 1,416 protein variants (mutated proteins) and their post-translationally modified forms, derived from 304 proteins. These have been used to annotate 532 complexes and 966 disease-specific reactions organized into 472 pathways and subpathways, and tagged with 349 Disease Ontology terms.
Tools and Data. Our services and software tools are designed for biologists, bioinformaticians, and software developers. Pathway data is available to view in our Pathway Browser, to analyze your own dataset, to download, and to access programmatically through our Content and Analysis Services. The ReactomeFIViz app for Cytoscape provides tools to find pathways and network patterns related to cancer and other types of diseases.
Documentation and Training. Visit our online User Guide to access documentation supporting pathway analysis of experimental data. The Developer's Zone provides detailed documentation regarding our software, tools, and web services. Training and learning materials can be found here.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information: If you have a question to ask or would like to give us your feedback, please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
As of Version 68 (March 2019), the RESTful API is deprecated and will be superceded by our ContentService. The ContentService is currently available through our website at https://reactome.org/ContentService/ and is designed for bioinformaticians, computer scientists, and software developers to access our pathway data. See more details about this API at https://reactome.org/dev/content-service.
If you have questions or concerns regarding this announcement, please email This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

Aiming to make our biological pathway knowledge more accessible, we've been offering for years the option to download the complete content of our knowledgebase as a PDF book. Following users feedback and requests, this book has now been updated to include yet more content in a freshly redesigned look and feel. Organised in 27 volumes, one per each top level pathway, the new version of the book includes our textbook-like lilustrations as well as regular-pathway and single-reaction diagrams.
In some cases the preferred option might be downloading part of Reactome's content. To do so we've included this option in our detail pages, the PathwayBrowser and the DiagramViewer. When downloaded from the PathwayBrowser, the resulting PDF will include the user's predefined colour profiles (diagram and analysis overlay) and will also include the analysis results when available. In all cases, resulting documents are fully navigable, including links to following and preceding events or any of the parent pathways in the hierarchy. Literarutre references include links to PubMed and every event has a link to its details page in Reactome's site.
The full book can be downloaded from our download section. For an example of the on-demand download please visit the Hemostasis details page or its view in the PathwayBrowser. All the above can be programmatically generated using this method from our ContentService.
- Details

Pathways in Reactome are curated in a generic cell and are agnostic to tissue types. However, different cell types have different functional requirements and consequently the underlying pathway activities also vary. Studying pathways in a tissue-specific manner will help to understand the biology in context.
Reactome now boasts a tool that can categorize pathways into different tissue types based on protein expression. We overlay tissue-specific protein expression data from the ExpressionAtlas database on empirically validated pathway information in Reactome. This facilitates the sorting of pathways from a generic cell to different tissue-types. This new feature allows users to select an experiment and analyse Reactome pathways in different tissues.
To try it out, select the "Tissue Distribution" tab, choose the tissues of your interest and click the "Go" button. The results are overlaid in the pathways overview and pathway diagrams similar to other analysis types. Users can cycle through the selected tissues via the small control panel displayed at the bottom of the viewport.
- Details
Up until now, we have placed existing regular pathway diagrams and textbook-style illustrations in our pathway detailed pages (e.g. Hemostasis, Platelet Adhesion to exposed collagen), leaving single reactions as the only type of events without a self-contained image. To fill the gap, an automatic algorithm to deterministically lay reactions out has been developed. It uses data directly from our graph-database to generate images without any kind of human intervention.
Long story short, the algorithm’s strategy places inputs on the left, outputs on the right, catalysts on top and regulators at the bottom. Each element is placed in its corresponding compartment, and these are nested following the Gene Ontology hierarchy. The algorithm supports normal and disease* reactions minimizing the space as well as avoiding lines to cross unnecessary elements in the display.
Finally, the reaction visualisation complies with the Systems Biology Graphical Notation (SBGN).
Some examples:
- Enzyme-bound ATP is released
- VCP-catalyzed ATP hydrolysis promotes the translocation of Hh-C into the cytosol
- Ubiquitination of PAK-2p34
- Defective ABCD1 does not transfer LCFAs from cytosol to peroxisomal matrix
*Disease reactions can either be classified as infectious, gain-of-function or lost-of-function. In the latter category some participants have to be crossed out.
- Details

New and Updated Pathways. In version V67, topics with new or revised pathways include Gene Expression (FOXO-mediated transcription), Immune Response (ROS, RNS production in phagocytes), Neuronal System (Activation of NMDA receptors and postsynaptic events), Programmed Cell Death (Apoptotic factor-mediated response), and Signal Transduction (GPCR downstream signaling and RHO GTPase activate NADPH oxidases).
New Illustrations. Illustrations with embedded navigation features are now available for ER to Golgi Anterograde Transport, Expression and Processing of Neurotrophins, and Signaling by NTRKs.
Thanks to our Contributors. Enrico Bertaggia, Timothy Donlon, Kasper Hansen, Oliver Nüsse, and Feng Yi are our external reviewers.
Annotation Statistics. Reactome comprises 12,788 human reactions organized into 2,256 pathways involving 11,066 proteins and modified forms of proteins encoded by 10,792 different human genes, 1,827 small molecules, and 155 drugs. These annotations are supported by 29,454 literature references. We have projected these reactions onto 139,298 orthologous proteins, creating 21,374 orthologous pathways in 18 non-human species. Version 67 has annotations for 1,564 protein variants (mutated proteins) and their post-translationally modified forms, derived from 299 proteins. These have been used to annotate 506 complexes and 962 disease-specific reactions organized into 467 pathways and subpathways, and tagged with 342 Disease Ontology terms.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

ReactomeFIViz is a Cytoscape app built upon Reactome pathways to help users perform pathway- and network-based data analysis and visualization. One of the most popular approaches to pathway analysis, which is an alternative to the traditional gene-list based pathway enrichment, is Gene Set Enrichment Analysis (GSEA). GSEA considers all genes with their scores based on a weighted Kolmogorov–Smirnov-like test and is a representative approach of second-generation pathway analysis. ReactomeFIViz implements features to perform GSEA analysis using Reactome pathways for a gene score file.
More details about the GSEA feature and the ReactomeFIViz app can be found here
- Details

Since version 57 we provide our data in a Neo4j graph database helping to reduce the complexity of the represented knowledgebase and allowing a more straightforward access to our content. Neo4j’s query language, Cypher, allows queries to be written in a more intuitive way and reduces the average response time per query by 93% (Fabregat et al., 2018).
The graph database also benefits Reactome in other aspects like (i) the creation of complex data quality assessment (QA) queries or (ii) supporting software that requires data pattern analysis, e.g. reactions classification. QA queries are executed during each quarterly release to identify instances that need to be checked out and corrected by Reactome curators ensuring that high-quality content is delivered to the final user. The reactions classifier project, for example, uses Cypher to formalise the concepts presented in Jupe et al. (2014) to generate a series of reports that help curators classify the reactions in Reactome.
The Content Service constitutes an easy API, based on the Representational State Transfer (REST) protocol, that provides access to the Reactome knowledgebase. It includes a set of methods classified in groups according to their functionality. For instance, expanding the pathways group reveals a set of methods that provide specific information about pathways such as the contained Events or the participating PhysicalEntities.
- Details
![]()
Textbook-like illustrations aim to improve the graphical representation of higher-level pathways in the Reactome events hierarchy, e.g., “signal transduction”, “apoptosis”, or “metabolism of proteins” whose pathway diagrams consisted of green boxes labeled with the names of sub-events, optionally located in cellular compartments and connected by arrows. These green-box diagrams feature limited navigation: clicking on a green box takes the user to that sub-event.
There was a general agreement that green-box diagrams are not that appealing, and put off users accustomed to textbook-quality illustrations of biological processes with striking, intuitively clear iconography. The project includes generation of scalable vector graphic (SVG) versions of illustrations, and a diagram module that makes these images interactive by enabling actions such as hovering over the items or selecting them to show the associated content in the details panel.
Developing this project has also driven the creation of the Icon Library; a consistent iconography compendium that ranges from simple protein labels to representations of organelles, receptors and cell types. The library has now been integrated in the main search as well as in the pages of their associated entities such as proteins or chemicals. Icons can be found by their name, description, designer and/or contributor.
The Icon Library is freely accessible (under a CC-BY 4.0 licence) and it is suitable for a broad range of purposes, from schematic pathway sketches in scientific presentations and publications to grant proposal illustrations. As the library was created to be a community resource, the invitation for third parties to contribute is still open. Aiming to achieve technical and artistic consistency, detailed guidelines are provided at https://reactome.org/icon-info. To acknowledge the community engagement, each icon is attributed to the author through a metadata file linking to a portfolio and/or ORCID id. As of September 2018, the library has considerably grown to 1,150 components.
- Details

As part of the International Open Access week, we would like to talk about another one of our open access network visualization and analysis tool – the Reactome FIViz app.
In order to improve our understanding of disease mechanisms and develop better personalized precision therapies for patients, many biological and clinical studies employ high-throughput techniques that generate large-scale data sets. Typically, these data sets are gene- or protein-based, and to better understand the relationships among interesting genes or proteins, researchers usually have to project them onto biological network contexts to provide holistic visualization and analysis platform for reducing the dimensionality of data using network modules.
To assist our users who would like to perform network-based analysis, we have constructed the Reactome Functional Interaction (FI) network which, covers 60% of the total human protein-coding genes, and was created by extracting interactions from manually curated pathways and predicting interactions based on a machine learning technique. We have developed the Cytoscape application (or app), called the “ReactomeFIViz” that uses this highly reliable FI network to support network-based data visualization and analysis. Users of our app can construct an FI subnetwork for a list of genes, perform network clustering to find network modules, annotate the subnetwork and modules, and perform survival analysis for network modules. Furthermore, the app can also perform pathway enrichment analysis using a gene score file, and pathway mathematical modeling based on probabilistic graphical models and Boolean networks. More documentation about the ReactomeFIViz app is available through our User Guide and the Cytoscape App Store.
- Details

Following up on the International Open Access week, we would like to remind our users that all our tools, including the analysis, are open access.
Pathway analysis methods have a broad range of applications in physiological and biomedical research. Our analysis suite currently implements an overrepresentation analysis, an expression data analysis and a species comparison tool. Using this service, users can submit their sample (list of identifiers) to get as result the most significants pathways. Results are overlaid in the different modules of our Pathway Browser and can be exported to different formats including a PDF report.
A light-weight client is integrated in our Pathway Browser. The tool suite is available via a RESTFul Web Service so all the available analysis tools can be easily integrated into third party software. More documentation for developers is available at our developer's zone.
- Details

The Systems Biology Graphical Notation (SBGN) project is an effort to standardise the graphical notation used in maps of biological processes. It aims to communicate biological knowledge more efficiently and accurately between different research communities in the life sciences.
Following this commitment, we’ve recently revamped our SBGN export methods and tools to provide more accurate representation of our pathway diagrams. Users have the option to either export a pathway diagram in SBGN through the PathwayBrowser, or get all human pathway diagrams from our downloads section.
- Details
Our PathwayBrowser now features advanced search capabilities powered by Solr to allow finding content throughout the whole knowledgebase. The user interface has been improved adapting to the findings of our last UX testing. The search within the Diagram Viewer widget enables users to define the scope of their search either limiting it to the content of the displayed diagram or expanding it to cover all pathways allowing our users to perform a search against all content without having to go the main search.
This new feature has also been enabled in our diagram and pathways overview widgets so third party web applications can already take advantage of it, allowing users to search Reactome content without abandoning the page they are in.
The new search features:
- Suggestions based on the introduced term.
- Listing the most recent searches.
- Scoping results to either the displayed diagram or the whole database.
- Filtering results by one or more entity types (i.e. Proteins, Chemical compounds, Reactions, etc.)
- Flagging a given entity to persist its highligthing.
To learn more, please check the Searching Reactome section in our User Guide.
- Details
New and Updated Pathways. In version 66, topics with new or revised pathways includeDisease (Loss of function of MECP2 in Rett syndrome and Defective Base Excision Repair Associated with MUTYH), Gene Expression (Transcriptional regulation by MECP2), Immune Response (OAS antiviral response), Metabolism of proteins (SUMOylation of DNA methylation proteins, SUMOylation of immune response proteins, SUMOylation of intracellular receptors, SUMOylation of SUMOylation proteins, SUMOylation of transcription cofactors, and SUMOylation of ubiquitinylation proteins), and Signal Transduction (Signaling by Erythropoietin).
Thanks to our Contributors. John Christodoulou, Rahul Krishnaraj, Kathy L McGraw, Marco Meras-Rios, Yusaku Nakabeppu, Einari Niskanen, Robert H Silverman, and Jürgen Wienands are our external reviewers.
New Ilustrations. Illustrations with embedded navigation features are now available for DNA replication and Protein localization.
Annotation Statistics. Reactome comprises 12,047 human reactions organized into 2,244 pathways involving 11,049 proteins encoded by 10,870 different human genes, 1,948 small molecules, and 11,823 complexes. New in this release are annotations of the functions of 139 drugs, 3 of them proteins and 136 small molecules. These annotations are supported by 28,829 literature references. We have projected these reactions onto 139,072 orthologous proteins, creating 21,283 orthologous pathways in 18 non-human species. Version 66 has annotations for 1,391 protein variants (mutated proteins) and their post-translationally modified forms, derived from 293 proteins, which have been used to annotate disease-specific complexes, reactions and pathways.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactomeare distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

We are pleased to announce that we are amongst the early adopters of the new Google Dataset Search.
Throughout the world, there are many thousands of open access data repositories hosted by public institutions, research projects, local and national governments, not-for-profit organisations, and many others. These online resources provide a variety of datasets, which are available in different formats and support many data standards. The Google Dataset Search engine enables easy access to these datasets, so that researchers, scientists, data journalists, data wranglers, or anyone else can quickly find the data for their work.
An example of how Reactome pathway data can be viewed in the Google DataSet Search is available at this link.
- Details

We now offer a new tool to download your analysis results as a single PDF file. This report contains a genome-wide overview, statistics for the most significant pathways and, for each significant pathway, it includes the corresponding diagram image with the analysis overlaid, a summation of the pathway, the related bibliography and the list of identifiers found.
When performing pathway analysis through our PathwayBrowser, you can download the report by clicking on the “Report (PDF)” button on the bottom-left corner of the Analysis tab. This feature is also available when implementing programmatic access to our AnalysisService through this method.
- Details
New and Updated Pathways. In version V65, topics with new or revised pathways include Immune System (Interleukin-9 signaling) and Signal Transduction (Signaling by NOTCH4 and Signaling by NTRK3 (TRKC)). Illustrations with embedded navigation features are now available for Carbohydrate Metabolism and Homology Directed Repair.
Thanks to our Contributors. Jorge Azevedo, Antonio Gómez-Outes, Jan Haavik, Eun-Kyeong Jo, Paula Licona-Limon, Jared Rutter, and Pantelis Tsoulfas, are our external reviewers.
Annotation Statistics. Reactome comprises 11,896 human reactions organized into 2,222 pathways involving 10,935 proteins encoded by 10,763 different human genes, 1,880 small molecules, and 11,674 complexes. These annotations are supported by 28,436 literature references. We have projected these reactions onto 159,163 orthologous proteins, creating 23,450 orthologous pathways in 18 non-human species. Version 65 has annotations for 1,339 protein variants (mutated proteins) and their post-translationally modified forms, derived from 289 proteins, which have been used to annotate disease-specific complexes, reactions and pathways.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence, A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
New and Updated Pathways. In version V64, topics with new or revised pathways include Signal Transduction (ESR-mediated signaling and Signaling by NTRK2), Metabolism (Biosynthesis of specialized proresolving mediators (SPMs)), and Metabolism of proteins (Peroxisomal protein import).
Thanks to our Contributors. Hanna Antila, Jorge Azevedo, Kumar Belani, Gerry Boss, Matthew Brenner, Eero Castrén, Arthur Cooper, Diana Downs, Marc Fransen, Trond Hansen, Hui-Chih Hung, Margaret James, Pidder Jansen-Duerr, Hideo Kimura, Luca Magnani, Steven Patterson, Paul Van Veldhoven, Alexander Weiss, and Herman Wolosker are our external reviewers.
Illustrations with embedded navigation features are now available for Digestion and absorption, DNA Double-Strand Break Repair, Signaling by receptor tyrosine kinases, Signaling by FGFR, MAPK family signaling cascades, MAPK1/MAPK3 signaling, Signaling by TGF-beta family, RHO GTPase Effectors, Signaling by Wnt, Signaling by Hedgehog, Death Receptor Signaling, and Intracellular signaling by second messengers.
Annotation Statistics. Reactome comprises 11,754 human reactions organized into 2,216 pathways involving 11,030 proteins encoded by 10,762 different human genes, 1,867 small molecules, and 11,561 complexes. These annotations are supported by 28,254 literature references. We have projected these reactions onto 140,720 orthologous proteins, creating 21,223 orthologous pathways in 18 non-human species. Version 64 has annotations for 1,339 protein variants (mutated proteins) and their post-translationally modified forms, derived from 289 proteins, which have been used to annotate disease-specific complexes, reactions and pathways.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence, A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
New and Updated Pathways. In version V63, topics with new or revised pathways include Immune System (Interleukin-20 family signaling, Interleukin-21 signaling, Interleukin-37 signaling, and other Interleukin signaling), Metabolism (Vitamin D (calciferol) metabolism), and Signal Transduction (Signaling by NOTCH3). Illustrations are now available for DNA Repair, Base Excision Repair, Signaling by GPCR, GPCR ligand binding, GPCR downstream signaling, and Translation.
Thanks to our Contributors. Laurence Bindoff, Roberta Carriero, Sandip Datta, Cecilia Garlanda, Elzbieta Glaser, Michael Holick, Tony Kouzarides , Alberto Mantovani, and Birgit Meldal are our external reviewers.
Annotation Statistics. Reactome comprises 11,426 human reactions organized into 2,179 pathways involving 10,996 proteins encoded by 10,739 different human genes, 1,764 small molecules, and 11,366 complexes. These annotations are supported by 27,694 literature references. We have projected these reactions onto 138,985 orthologous proteins, creating 20,932 orthologous pathways in 18 non-human species. Version 63 has annotations for 1,334 protein variants (mutated proteins) and their post-translationally modified forms, derived from 287 proteins. These have been used to annotate 506 disease-specific complexes and 906 disease-specific reactions organized into 453 pathways and subpathways, and tagged with 294 Disease Ontology terms.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome annotation files and interaction data derived from Reactome are distributed under a Creative Commons Public Domain (CC0 1.0 Universal) Licence,. A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will apply to all software and code, database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials. A full description of the new and updated content is available on the ,..
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

Since our inception, we have been an open source and open access resource, free for use by anyone under the terms of a Creative Commons Attribution 4.0 International (CC BY 4.0) license. This license granted parties the non-exclusive right to use, distribute and create derivative works based on Reactome, provided that the works are correctly attributed to OICR, NYUMC, EBI, and OHSU.
In line with the growing movement to provide free open data in the public domain, and to better support the needs of our user community, we are updating the licensing agreement for some of our web content and data to reflect the Creative Commons Public Domain (CC0). CC0 is the "no copyright reserved" option in the Creative Commons toolkit. It effectively means relinquishing all copyright and similar rights that we hold in a work and dedicating those rights to the public domain.
A Creative Commons Public Domain (CC0 1.0 Universal) Licence will now cover all Reactome annotation files, e.g. identifier mapping, specialized data files, and interaction data derived from Reactome.
A Creative Commons Attribution 4.0 International (CC BY 4.0) Licence will continue to apply to all software and code, e.g. relating to the functionality of the reactome.org, derived websites and webservices, the Curator Tool, the Functional Interaction application, SQL and Graph Database data dumps, and Pathway Illustrations (Enhanced High-Level Diagrams), Icon Library, Art and Branding Materials.
For information about how to properly credit data use, please review the Reactome License and the Creative Commons FAQ, or contact the This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details

A new research article titled “The Reactome Pathway Knowledgebase” has been published in the forthcoming 2018 NAR Database Issue. The paper describes the deployment of a Neo4J graph database on our production server, development of a new high-performance in-memory implementation of our overrepresentation data analysis tool, improvements to the Pathway Diagram Viewer, and implementation of the new Enhanced High Level Diagrams (EHLDs). More publications from the Reactome Team can be found here.
- Details

As part of the ongoing evolution of our website, we are proud to announce the launch of our new logo. Reactome has grown and evolved over the last 14 years, and we felt it was time for a change.
The new logo brings to the forefront the value and quality of the information and analysis tools that can be found in our curated database of pathways. The layering in our logo highlights the transparency of Reactome’s nature while the rounded typography matches our openness.
Derived from a shape that exists in nature and at the same time gives structure, the logo evokes the idea of discovering by unwrapping the layers of knowledge that surround biological events.
We have refreshed our logo to reflect who we are today and to symbolize our dynamic future. Rest assured though: our high-quality data and services, and our dedication to you will remain the same!
For more information, please go to Our Logo.
- Details

We’ve launched our new website and are excited to introduce you to our new look! Reactome is inviting its users to explore its new website. The new website has been designed to provide the ultimate user-friendly experience with improved navigation and functionality throughout. Created with the user experience firmly in mind, the new web interface has been designed using the latest technology, so the site is compatible with today's browsers and mobile devices. The site includes extensive documentation to help users understand Reactome’s complete range of tools for viewing pathway diagrams, analyzing experimental data and exploring protein-protein interaction networks. For software developers, our technical documentation and use cases provide a detailed overview of our extensive web services, to access our curated pathway data (ContentService) and analytical tools (AnalysisService). Biologists and bioinformaticians can now benefit from richer online content that is easier to navigate and share with others, assisting with pathway data analysis and visualization.
- Details
New and Updated Pathways. In version V62, topics with new or revised pathways include Developmental biology (Signaling by Robo receptor), Gene expression (Transcriptional regulation by E2F6 and Transcriptional regulation by RUNX2), and Immune System (Interleukin-7 family signaling, Interleukin-15 family signaling, Interleukin-35 family signaling, and Interleukin-38 family signaling). Illustrations are now available for Aquaporin-mediated transport, Cellular senescence, Epigenetic regulation of gene expression, Gene Expression, Negative epigenetic regulation of rRNA expression, O2/CO2 exchange in erythrocytes, Peptide hormone metabolism, Positive epigenetic regulation of rRNA expression, Post-translational protein modification, Response to metal ions, Signal Transduction, and SLC-mediated transmembrane transport.
Thanks to our Contributors. Patricia Ducy is our external author. Patricia Ducy, Jorg Goronzy, Anna Herlihy, Alexander Jaworski, Umesh Kumar, Javier Francisco Mora, Manoj Patidar, Yuliya Pylayeva-Gupta, Kailash Singh, and Ren Sun are our external reviewers.
Annotation Statistics. Reactome comprises 11,302 human reactions organized into 2,176 pathways involving 10,878 proteins encoded by 10,712 different human genes, 1,768 small molecules, and 11,284 complexes. These annotations are supported by 27,526 literature references. We have projected these reactions onto 121,709 orthologous proteins, creating 20,854 orthologous pathways in 18 non-human species. Version 62 has annotations for 1,334 protein variants (mutated proteins) and their post-translationally modified forms, derived from 285 proteins. These have been used to annotate 506 disease-specific complexes and 906 disease-specific reactions organized into 453 pathways and subpathways, and tagged with 294 Disease Ontology terms.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
As part of our efforts to give our modeling user community better experience, we have updated the SBML export to Level 3 Version 1, which is organized in a modular manner. Our initial export provides a richer annotation syntax and we will explore supporting other SBML Level 3 Packages in the future. The SBML data export can be used by any tool that supports SBML L3V1.
The SBML Level 3 Version 1 export is available for download here: https://reactome.org/download/current/homo_sapiens.3.1.sbml.tgz.
We are also providing a programmatic interface to access these updated SBML files through our ContentService for all species at https://reactome.org/ContentService/#!/exporter/toSBMLUsingGET
Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University Langone Medical Center, Oregon Health and Science University, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
- Details

The Reactome team has released new versions of our protein-protein interaction files derived from reactions and complexes. These files were updated following user feedback, with the goal of providing extra annotation features through support from the PSI-MITAB 2.7 data format. Interactions are computationally generated based on the data stored on complexes and reactions. The interactions provided by Reactome are not curated and are not experimental data. In addition, the complexes and reactions in species other than human are derived by orthology inference from the corresponding human complexes and reactions. Tab-delimited formatted files are also provided for human and all species.
The four new files are:
- Full list of protein-protein interactions, incl. non-human species [PSI-MITAB]
- Full list of protein-protein interactions, incl. non-human species [tab-delimited]
- Human protein-protein interactions [PSI-MITAB]
- Human protein-protein interactions [tab-delimited].
Documentation about these files is available from the Data Download page.
The former versions of the protein-protein interaction files are still available but will be removed from service as of Version 62 in September 2017. We encourage users that programmatically access these files to update the scripts.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University Langone Medical Center, Oregon Health and Science University, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
New and Updated Pathways. In version V61, topics with new or revised pathways include: Gene expression (Transcriptional regulation by RUNX1), Immune System (Interleukin-12 family signaling), Signal Transduction (PTEN Regulation), and Transport of small molecules (Intracellular oxygen transport).
Thanks to our Contributors. Arkaitz Carracedo and Leonardo Salmena were our external authors. Sabine Bailly, Thorsten Burmester, Linda Shyue Huey Chuang, Yoshiaki Ito, Nisha Kriplani, Nick Leslie, and Esther van de Vosse were our external reviewers.
Annotation Statistics. Reactome comprises 11,042 human reactions organized into 2,148 pathways involving 10,940 proteins encoded by 10,691 different human genes, 1,763 small molecules, and 11,041 complexes. These annotations are supported by 26,859 literature references. We have projected these reactions onto 120,804 orthologous proteins, creating 20,780 orthologous pathways in 18 non-human species. Version 61 has annotations for 1,334 protein variants (mutated proteins) and their post-translationally modified forms, derived from 285 proteins. These have been used to annotate 506 disease-specific complexes and 906 disease-specific reactions organized into 453 pathways and subpathways, and tagged with 294 Disease Ontology terms.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
New and Updated Pathways. In version V60, topics with new or revised pathways include: Cell cycle (Cyclin D associated events in G1), Cell-cell communication (SDK interactions), Cellular response to external stimuli (HSP90 chaperone cycle for steroid hormone receptors (SHR)), Disease (Diseases of Mismatch Repair), Gene Expression (TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest and Transcriptional Regulation by the CBFB:RUNX3 complex), Immune System (Butyrophilins and Regulation of complement cascade), Metabolism (Synthesis of IP2, IP, and Ins in the cytosol, Synthesis of PIPs at the early endosome membrane, Synthesis of PIPs at the ER membrane, Synthesis of PIPs at the late endosome membrane, and Synthesis of PIPs at the plasma membrane), Metabolism of proteins (CREB3 factors activate genes, Neddylation, Protein ubiquitination, and SUMOylation of chromatin organizing proteins), Mitophagy (Receptor Mediated Mitophagy), Neuronal System (Receptor protein tyrosine phosphatases interactions), Organelle biogenesis and maintenance (Cristae formation), and Transport of small molecules (Mitochondrial calcium ion transport).
Thanks to our Contributors. Wei-Chih Yang and Jian Lu are our external authors. Joseph Ainscough, Sanjeevani Arora, Jorge E Azevedo, Jeehyeon Bae, Lucia Banci, Gautam Bhave, David R Brown, Roberta Bulla, Alexandre M Carmo, Linda Shyue Huey Chuang, Karlene A Cimprich, Laura Crisponi, Alain de Bruin, Luisa Di Stefano , Ilaria Drago, Pablo C Echeverria, Du Feng, Emer S Ferro, Dianne Ford, Frances V Fuller-Pace, Cem Gabay, Dominique Gagliardi, Marcia Haigis, J Wade Harper, Barry Honig, Yoshiaki Ito, Veerle Janssens, Nathalie Josso, Jaewon Ko , Vera Kozjak-Pavlovic, Anastasia Kralli, Paul J Lehner, Dominique Leprince, Bruce D Levy, Wei Li, Francisco Lozano, Jian Lu, Michael J Matunis, Birgit Meldal, Violaine Moreau, Kyungjae Myung, Joseph H Neale, Christian Obinger, R Jeroen Pasterkamp, Richard Phipps, Didier Picard, Elah Pick, David A Rhodes, Pier e P Roger, Mark G Rush, Joshua R Sanes, Martin Schröder, Pierre Thibault, Dick J H van den Boomen , Thomas E Van Dyke, Roberto M Vanacore, Nobutaka Wakamiya, Qinglu Wang, Bart Westendorp, Sandra E Wiley, Miriam Wittmann, Guilherme Xavier, Wei-Chih Yang, Ali A Zarrin, and Bing Zhu are our external reviewers.
Annotation Statistics. Reactome comprises 10,754 human reactions organized into 2,132 pathways involving 10,907 proteins encoded by 10,658 different human genes, 1,763 small molecules, and 10,748 complexes. These annotations are supported by 26,384 literature references. We have projected these reactions onto 115,881 orthologous proteins, creating 20,701 orthologous pathways in 18 non-human species. Version 60 has annotations for 1,503 protein variants (mutated proteins) and their post-translationally modified forms, derived from 285 proteins. These have been used to annotate 506 disease-specific complexes and 906 disease-specific reactions organized into 453 pathways and subpathways, and tagged with 294 Disease Ontology terms.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, Oregon Health and Science University, New York University Langone Medical Center, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
A new Reactome paper titled “Reactome pathway analysis: a high-performance in-memory approach” has been published in BMC Bioinformatics. More publications from the Reactome Team can be found here.
- Details
A new Reactome paper titled “Functional Interaction Network Construction and Analysis for Disease Discovery” has been published in Methods in Molecular Biology. More publications from the Reactome Team can be found here.
- Details
New and Updated Pathways. In version V59, topics with new or revised pathways include: Disease (Listeria monocytogenes entry into host cells), Hemostasis (Cell surface interaction at the vascular wall), Immune System (Butyrophilins, Interleukin 10 signalling, and Interleukin-4 and 13 signaling), Metabolism (Nicotinate metabolism, Synthesis of PIPs at the nuclear envelope, Vitamin B5 (pantothenate) metabolism, and Aryl hydrocarbon receptor signalling), Metabolism of proteins (E3 ubiquitin ligases ubiquitinate target proteins, Peptide-ligand binding receptors, Protein methylation, RAB geranylgeranylation), Signal transduction (Class A/1 (Rhodopsin-like receptors), and Vesicle-mediated transport (TBC RABGAPs).
Thanks to our Contributors. Our external reviewers are Jorge Azevedo, Ester Boix, Lu Deng, Pål Falnes, Vardan Karamyan, Samuel Leibovich, Weei-Chin Lin, Charuta Palsuledesai, Joel Pomerantz, Walter Reith, Sylvie Ricard-Blum, Christian Schwerk, Bruce Spiegelman, and Xiaochun Yu.
Annotation Statistics. Reactome comprises 10,391 human reactions organized into 2,080 pathways involving 10,624 proteins encoded by 10,381 different human genes, and 1,735 small molecules. These annotations are supported by 25,449 literature references. We have projected these reactions onto 115,881 orthologous proteins, creating 20,164 orthologous pathways in 18 non-human species. Version 59 has annotations for 1,496 protein variants (mutated proteins) and their post-translationally modified forms, derived from 285 proteins. These have been used to annotate 506 disease-specific complexes and 897 disease-specific reactions organized into 447 pathways and subpathways, and tagged with 294 Disease Ontology terms.
About the Reactome Project. Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, New York University Medical Center, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information. Please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
The Reactome team is pleased to announce that it met a major milestone in October 2016 with the annotation and release of its 10,000th human protein. Reactome (www.reactome.org) is an open access curated knowledgebase which relates human genes, proteins and other biomolecules to the biological pathways and processes in which they participate. It is a key resource for the biomedical research community, and is widely used by researchers around the world to interpret high-throughput experiments in genetics, genomics and proteomics. Given that the human genome contains roughly 20,000 protein-coding genes in total, the annotation of the 10,000th protein means that Reactome now covers half of the protein-coding portion of the genome. This makes Reactome the most comprehensive open access pathway knowledgebase available to the scientific community.
By relating genes and proteins to normal and abnormal biological pathways, Reactome allows researchers to identify patterns in large data sets. For example, researchers can use Reactome to reduce an experiment that identified thousands of genes whose activities are altered in a disease to a manageable number of key biological pathways that are disrupted by these changes. Researchers can then combine Reactome with other databases to find drugs and protein targets that might reverse the pathway alterations, or to devise ways of diagnosing the disease at an early stage. Via its web site, online tools, and specialized visualization and analysis applications, Reactome has been incorporated into more than 400 third-party genome analysis tools, and has been cited more than 4,000 times in the scientific literature.
Reactome has been in continuous operation since 2004 and is an international collaboration among the Ontario Institute for Cancer Research in Canada, New York University School of Medicine in the United States, and the European Bioinformatics Institute in the United Kingdom. It is staffed by expert biological curators, bioinformaticians and computer scientists. Much of its content is provided by community authors and peer reviewers who are assisted by the curatorial staff. The Reactome content, including pathway data and the software infrastructure, are available to all comers free of charge under a Creative Commons open access license. Reactome is supported by grants from the US National Institutes of Health, the Ontario Research Fund, the University of Toronto, OpenTargets, Genome Canada, and the European Molecular Biology Laboratory.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
New and Updated Pathways. With version V58, Reactome has annotations for over 10,000 human proteins. New or revised pathways include: Cell cycle (FBXL7 down-regulates AURKA during mitotic entry and in early mitosis), Developmental biology (Keratinization), Disease (Oncogenic MAPK signaling and Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer’s disease models), Gene expression (PI5P Regulates TP53 Acetylation, Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors), Immune system (Neutrophil degranulation and Antimicrobial peptides), Metabolism of proteins (Synthesis of active ubiquitin: roles of E1 and E2 enzymes), Signal transduction (Signaling by MET and Downregulation of ERBB2 signaling), and Vesicle-mediated transport (RAB GEFs exchange GTP for GDP on RABs).
A pathway illustration is available for Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer’s disease .
Thanks to our Contributors. Our external author is Kavita Shah. Emily Ayoub, Jorge Azevedo, Walter Birchmeier, Miroslav Blumenberg, Maria Bogachek, Nullin Divecha, Roman Dziarski, Rhys Grant, David Hains, Niels Heegard, Guustaaf Heynen, Catherine Lindon, Andrea Marat, Robert Stephens, Michel Tremblay, and Ronald Weigel are our external reviewers.
Reactome comprises 10,168 human reactions organized into 2,069 pathways involving 10,461 proteins encoded by 10,221 different human genes, and 1,710 small molecules. These annotations are supported by 24,974 literature references. We have projected these reactions onto 110,710 orthologous proteins, creating 19,991 orthologous pathways in 18 non-human species.
Reactome is a collaboration between groups at the Ontario Institute for Cancer Research, New York University Medical Center, and The European Bioinformatics Institute. Reactome data and software are distributed under the terms of the Creative Commons Attribution 4.0 License. A full description of the new and updated content is available on the Reactome website.
Follow us on Twitter: @reactome to get frequent updates about new and updated pathways, feature updates, and more!
For more information please contact our This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
In version V57, topics with new or revised pathways include: Developmental biology (RET signaling), Disease (Signaling by FGFR in disease and Defective CFTR causes cystic fibrosis), Gene expression (rRNA modification in the mitochondrion and Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors), Immune system (ER-Phagosome pathway, Interleukin-7 signaling, Regulation by endogenous TLR ligand, and TCR signaling), Metabolism (Lipid digestion, mobilization, and transport and Phosphate bond hydrolysis by NTPDase proteins). Metabolism of proteins (Deubiquitination), Neuronal system (Interactions of neurexins and neuroligins at synapses and SALM protein interactions at synapse). Signal transduction (EGFR downregulation, Signaling by FGFR1, and FGFRL1 modulation of FGFR1 signaling), Transmembrane transport of small molecules (ABC-family proteins mediated transport) and Vesicle-mediated transport (Clathrin-mediated endocytosis).
Our external author is Alba Sanchis. Costin Antonescu, John Bergeron, Daniel Bogenhagen, Nunzio Bottini, Guang-Chao Chen, Igor Dawid, Regina Fluhrer, Noriko Gotoh, Francesca Granucci, Richard Grose, Paul Heppenstall, Jing Hu, Jan Huertas, Wenqin Luo, Malay Mandal, Birgit Meldal, Daniel Morales, Tatsunori Nishimura, Ronald Petralia, Gail Seabold, Sunny Sharma, Stephanie Stanford, Jean Sévigny, Philip Washbourne, Ivan Zanoni, and Valeria Zarelli are our external reviewers.
© 2024 Reactome







